I am an Evolutionary Biologist with research line intersecting the fields of zoology, ecology, evolution and conservation of non-model organisms, particularly arthropods. The main goals of my research are to understand broad scale morphological and molecular evolution, global biogeography, molecular ecology, and face the biodiversity crisis through taxonomy and conservation. Specifically, I aim to answer the following questions:
- How many species of arthropods are there?
- How do species relate to each other and how they got to where they are in the world?
- How do species originate and persist through evolutionary time?
- What biotic and abiotic factors interfere in species distribution?
To tackle these questions, I make use of an interdisciplinary approach that integrates systematics, biogeography, genomics, bioinformatics and ecology. Details on ongoing projects below. List of publications here.
Ongoing projects
|
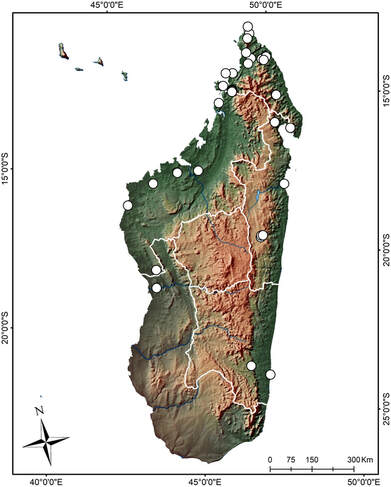
Microendemics of Madagascar: diversity, systematics and phylogeography of Madagascan Schizomida based on morphology and ultraconserved elements (UCEs)
Schizomids are small arachnids that live in the leaf litter of tropical and subtropical forests and inside caves. They are short-range endemics and low dispersal organisms, making them excellent candidate models for biogeography studies. Little is known about schizomid biogeography, diversity, and evolution, and no previous molecular work has been done on Madagascan schizomids. We aim to bridge this gap by investigating the taxonomy, systematics, and phylogeography of Madagascan schizomids using UCE data from museum-preserved and newly collected material. Specifically, we propose to [1] describe new species and genera of schizomids from Madagascar, [2] reconstruct their phylogenetic histories, and [3] analyze patterns of gene flow, morphological variability, and species diversification in a phylogeographic context. We will also collect schizomid specimens in California, which will serve as outgroups in our phylogenetic and phylogeographic analyses. The results of our project have the potential to provide valuable insight into the systematics of a little-studied arachnid group from an under-sampled region of the world. This project will also generate the first ultraconserved element sequences and GGBN records for schizomids and the first quality samples of Schizomida in the biorepository.
|
|
Funding:
Additional support:
|
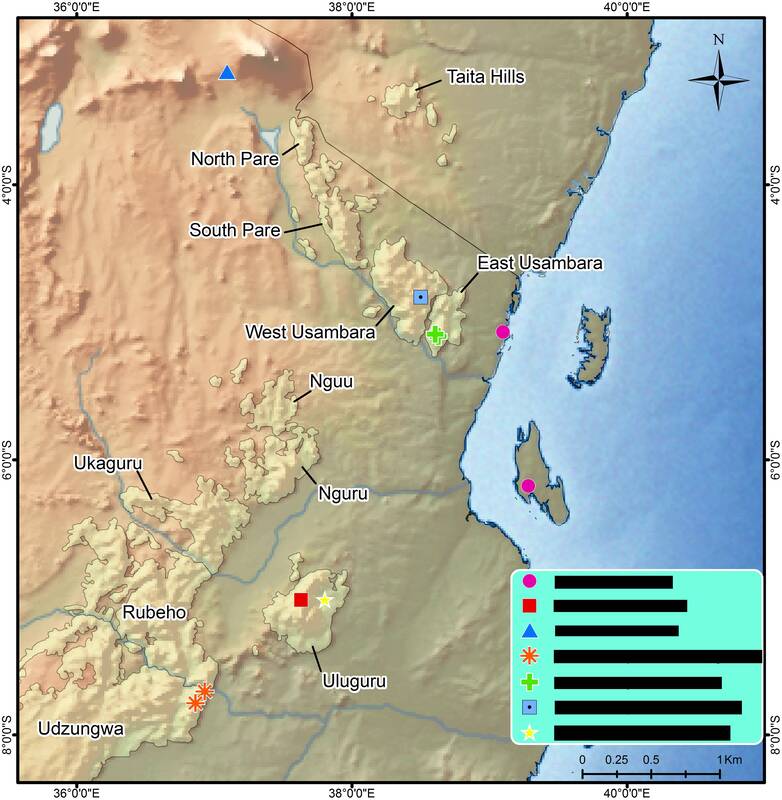
Enhancing the knowledge on Afromontane biodiversity hotspots: phylogenomics, biogeography and the GGBN database
The distinct shape and exclusive biota of mountains make these environments good models for understanding biogeography and diversification processes. Afromontane areas, in particular, are of high interest due to their having a great number of endemic species often with ancient ages, but the origin of this high diversity is still debated. Traditionally, taxa on African mountains have been considered relictual, resulting from historical glacial cycles that forced taxa to move up into montane refugia. Alternatively, persistent orographic rain and mist are suggested as being the causes for high diversity in African mountaintops, which serve not only as refugia (for the relictuals) but also as centers of speciation. Among African highlands, the Eastern Arc Mountains (Tanzania and Kenya) are a hotspot of biodiversity and, due to their old age and geographical position, present an interesting opportunity to examine diversification processes. The knowledge on the diversification of most animals and plants in these mountains, however, is still incipient, especially for low-dispersing, understudied arthropods. To address this gap in knowledge, this project aims to study the biogeography and evolution of the endemic fauna on the Eastern Arc mountains using four low-dispersal, endemic lineages of spiders: Cyatholipidae, Linyphiidae, Palpimanidae, and Phyxelididae. The project goal is to sequence hundreds of genomic ultraconserved elements (UCEs), build new phylogenies for the families and analyze the relationships of the Eastern Arc species compared to other close relatives distributed worldwide. Additionally, the phylogenies will be time-calibrated and a biogeographic analysis will be performed to investigate patterns and processes of dispersal/vicariance in the Eastern Arcs. This project also aims to publicly release in GGBN the great spider collection from the Eastern Arc Mountains deposited in the Natural History Museum of Denmark. |
|
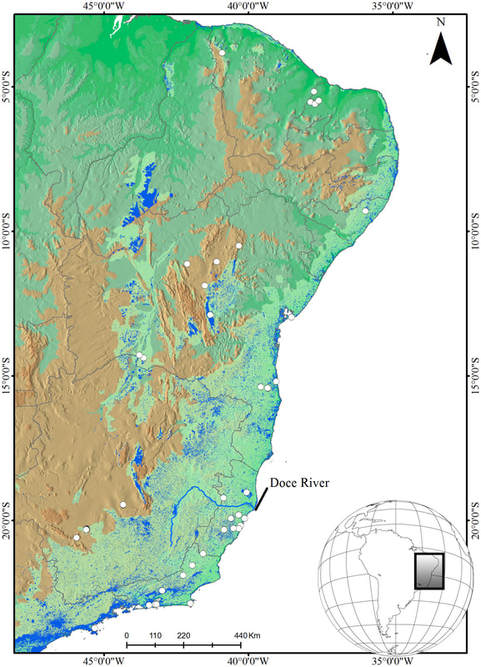
Phylogeography of the Atlantic Forest using whip spiders as a model organism
This project aims to use next generation sequencing methods and a multi-locus approach to analyze the genome of an invertebrate group, the whip spiders of the genus Charinus. Expanding on the work done for my doctoral thesis, the plan is to collect fresh material from 20 points in the Atlantic Forest, from the northernmost to the southern region of the forest, extract, amplify and sequence the DNA, and analyze the population genetics of the species sampled. With this fine-scale approach to the forest diversity, it will be possible to model species distributions, understand past effects of tectonic events and climatic changes in the forest's Charinus fauna, and predict the effects of future climate changes in this hyper diverse forest. This will be one of the first phylogeographies of the AF using an invertebrate as a model, the first phylogeography using genomic data. This research is an important step towards understanding South American biodiversity, and the broad question of why some areas of the planet harbor a greater degree of diversity than others. |
|
Funding:
|
Additional support:
|
|
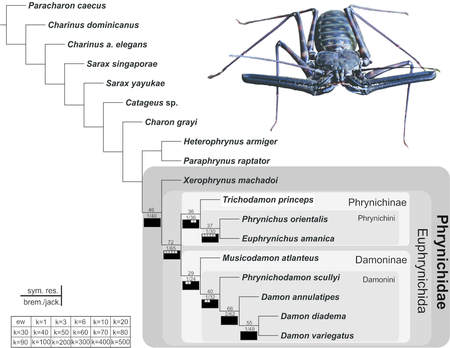
Systematics, taxonomy and biogeography of Amblypygi
The Amblypygi is an amazing group of organisms that has much of its taxonomy and systematics unresolved. This project has the aim of addressing questions relating to evolution of these animals, their conservation and relationship. To reach that, I use morphology and DNA (multilocus data [the usual suspects] and Ultraconserved Elements [UCE]). To explore the historical and ecological biogeography of the group, I date calibrate molecular trees (using BEAST), do ancestral state reconstruction (with BioGeoBEARS), and study species limits and genetic diversity (population genetics and phylogeography using a pipeline for SNPs from UCE data). |
Support:
|
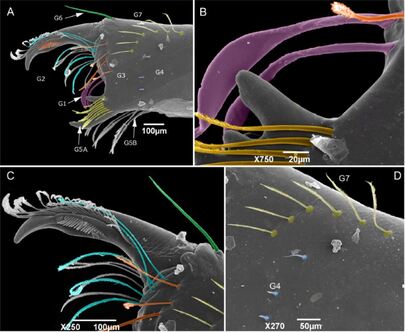
Systematics and taxonomy of Schizomida
The poorly known arachnids order Schizomida has its diversity underestimated, a possible result of its cryptic habitat and small size. This project is trying to resolve the order's taxonomy and phylogeny with the description of new species and raising of new characters for a morphological phylogeny of the group. Support:
|